Living
in the
brave
new
genomic
era

If you’re reading this page, you’re walking around with a human genome.
The human genome is the full collection of DNA that allows for a human life.
While all humans are encoded by the human genome, each one of us carries a slightly different version. This small variation in DNA sequence helps explain our unique traits (height, eye color, food preference.)
The first human genome was sequenced in 2003. In the following 17 years, we’ve seen incredible developments with DNA based technology. Individuals can now decode their genome and gain insight into their ancestry, lifestyle and general health.
Perhaps most significantly, scientists are learning to correct genetic mistakes in the human genome.
In 2017, we witnessed the first FDA approval for a gene therapy. An inherited form of blindness can now be treated with a genetic fix.
This web page serves as a platform to educate the general public about our new genomic era.
For each phase of life, I review different ways we genetic information and services can be utilized.
Click on a topic below to jump straight to that section.
Living in the Genomic Era
To keep up with this fast moving field, I’ll update this page every 6 months.
Please share the page by clicking icons below.
The year is 2020.
We now know the human body is an intricate machine that receives its marching orders from a long string of chemicals called DNA.
We know that DNA dictates the construction of our physical body. As we move through life, our DNA code continues to instruct and influence our health, our emotions, our behavior.
What is the human genome and how does it work?
The human genome is a huge database made from 3 billion pieces of linear code. This code, called DNA, consists of 4 different chemicals (Adenine, Guanine, Cytosine and Thymine). We store this DNA code inside the cells of our body. Our cells refer to the code when we need to construct new biological material for our body.
Approximately 20,000 genes are packaged within the human genome.
These genes are the segments of our DNA code that translate into the new biological material in our body.
When necessary, cells in our body convert our linear genes into sophisticated, three-dimensional proteins. This complex shape allows proteins to perform complex biological work in our body.
When all this work is performed correctly, a human being remains alive and healthy.
As mentioned above, each one of us is walking around with a unique version of the human genome. Our unique genomes are a combination of our parents’ genomes.
The genetic code for all humans is about 99.9% identical.
It’s the 0.1% difference that explains much of our individual nature.

artwork: Alberto Giacometti
Human figures
Genetic testing
Understanding the DNA sequence of our genes helps explain how our body works.
If we carry the correct DNA sequence for a gene, then we can produce the correct shape for the corresponding protein. A properly shaped protein performs its biological job properly.
In contrast, if we carry an incorrect DNA sequence for a gene, we produce an oddly shaped protein for that gene. This oddly shaped protein may fail to perform its biological job and, therefore, cause a related health issue.
When we test our DNA, we are determining the exact sequence of letters (A,C,G,T) in our genes.
This genetic information provides insight into our ancestry, our lifestyle and our general health.
If you suffer from an un-diagnosed disease, a genetic test may also determine the genetic basis for that disease. This knowledge could lead to a precise treatment for your illness.
Below, I discuss 3 different types of genetic testing.
- direct to consumer genetic testing (learn how your DNA relates to ancestry/health/wellness)
- test your whole genome with a counselor (seek out a genetic basis for a disease)
- biopsy: sequence DNA from tumor cells (search for mutations that may point to a targeted therapy)
Click a topic above to jump straight to that section.
Direct to consumer genetic testing
The most popular form of DNA testing is called direct to consumer genetic testing. If you haven’t already sent in your DNA for testing, you’ve likely seen advertisements for ancestryDNA, LivingDNA or 23andMe.
A handful of companies are competing for this retail market. Some companies specialize in ancestry data while others focus on health and wellness information.
In general, here’s how direct to consumer genetic testing works:
- Customer purchases the testing service online (cost between $70-200).
- DNA collection kit is mailed to customer.
- Customer spits in a tube and mails tube to genetic testing company (saliva contains buccal cells).
- Company collects buccal cells from saliva and isolates DNA from these cells.
- DNA is sequenced and genetic information is digitally sent to customer.
Direct to consumer genetic testing has spiked in the past few years. An inflection point in sales began in 2016.
In 2017, the number of people who analyzed their DNA doubled. This number began exceeding 12 million individuals.
Estimates now suggest that 1 in 25 American adults have access to their personal genetic information. This is incredible considering the very first human genome was completely sequenced in 2003.
It’s important to know that direct to consumer genetic tests only explore a small portion of your entire genome.
Why would you only sequence a portion of your genes?
Why not sequence all 20,000 genes?
Here is the most simple answer. The science community only understands the function of a small portion of the human genome.
Of the 20,000 human genes, we only really understand about 200 genes. Therefore, direct to consumer genetic companies use gene chips to only collect DNA sequence from the few genetic regions that are informative in regards to ancestry, lifestyle or health.
artwork: Ryan Kitson
On the table below, I list examples of genes that we do understand.
Scientific studies have determined how variations in the DNA sequence of these genes will affect the human body.
Direct to consumer genetic tests can determine your specific DNA sequence for these genes.
| Gene | Genetic variation that affects health or lifestyle |
|---|---|
| APOE | Individuals with the E4 version of APOE may experience an increased risk of developing late-onset Alzheimer’s disease. |
| CYP1A2 | If a region of your CYP1A2 gene contains a T (thymine) instead of a C (cytosine), then you metabolize caffeine more aggressively and likely drink more coffee. |
| ALDH2 | If a region of your ALDH2 gene contains a G (guanine), then you don’t process alcohol efficiently and your face may blush red when drinking alcohol. |
Keep in mind, each gene in our body may perform multiple functions. While we understand some function of these genes, we still don’t understand everything they’re doing in the human body.
Sequence your entire genome
Some people need a more extensive genetic analysis than a direct to consumer test can provide.
If you’re suffering from an un-diagnosed illness or have a family history of an inherited disease, then you may want to consider sequencing your entire genome.
Most direct to consumer tests will only sequence a handful of genes, however, a whole genome test will sequence your entire genome (about 20,000 genes). That’s a lot of genetic information.
Currently, the most common form of a whole genome test is called whole exome sequencing. This test will determine your unique sequence for all of the protein coding genes within your genome.
Whole exome sequencing may cost between 5-10K. This cost should include consultation and the interpretation of results with a genetic counselor.
Individuals with cardiovascular disease or cancer could consider using whole exome sequencing to seek additional insight into their condition (1).
This video explains how whole exome sequencing tests all the genes within our genome.
The video is also helpful if you’re unclear about DNA mutations or the difference between genes and genomes.
What are the possible benefits of whole exome sequencing?
- a genetic diagnosis for a sick individual could guide medical care
- diagnosis may determine the heritability of the condition (i.e. will this be passed on to kids?)
- information could allow a healthy person to optimize their life (not likely at this time)
At this point in time, whole exome sequencing is mostly intended for children with health issues or adults with an un-diagnosed disease. In contrast, a somewhat normal and healthy adult is unlikely to benefit in a meaningful way from whole exome sequencing.
While current technology allows us to sequence the entire human genome, doing so for healthy people currently has no clear health benefit. Simply because it is possible to sequence entire human genomes does not mean its broad application in the healthy is a good idea – at least not yet.
As mentioned above, we currently only understand a small portion of the human genome. About 90% of human genes remain fairly mysterious to scientists. We simply don’t know exactly how these genes function in the human body. Therefore, DNA information from these genes will be less valuable (and less actionable) for a patient.
Alternatively…
- You can sequence only a panel of genes that are known to relate to your particular illness (breast/ovarian cancer, colon cancer, dilated cardiomyopathy, etc.)
- You can take a panel test called ACMG59. This test screens for variants in 59 actionable genes. These genes relate to cancer, cardiovascular disease and some metabolic conditions. These gene variants are highly penetrant and actionable. They’re actionable because there’s an intervention associated with the genes. If you test positive for one gene variant, you can then take steps to prevent the associated disease or improve diagnosis.
- You can also choose to sequence your entire genome, all 3 billion bases. A true whole genome sequence would decode 100% of your DNA, this includes coding DNA and non-coding DNA. Whole exome sequencing only decodes about 1.5% of your genome, it ignores non-coding DNA. Sequencing the entire genome rarely delivers more value than whole exome sequencing. In the future, as we unlock more connections between DNA sequence and human health, the 100% genome sequence will become more common.
Speak with your physician or a genetic counselor to help decide which genome test is best for you.
Test DNA from cancer cells
The genome we inherit from our parents does not remain static throughout our life. As we age, we accumulate errors in the DNA sequence of our genome.
Our body is a machine but like most machines – our body is an imperfect machine.
For example, throughout our lifetime, we accumulate single base mutations. An Adenine may pop out of sequence on a gene and be replaced with a Thymine. This definitely changes the DNA sequence for that gene and it may render the corresponding protein useless.
Sometimes a large chunk of a gene will break off, leaving the protein completely useless. Mistakes happen.
These mistakes are called mutations. Many of our accumulated mutations will have no effect on our health. However, sometimes a DNA mutation occurs in a sensitive location in our genomes. If a mutation lands in the middle of an important gene, it can disrupt the function of that gene and possibly cause a disease.
If you smoke a pack of cigarettes every day, then your lung cells will be bombarded with carcinogens (cancer causing chemicals). These chemicals will heavily mutate the DNA within your lung cells. An accumulation of DNA mutations in lung cells will eventually push these cells to become the cancerous versions of healthy lung cells. This is how lung cancer begins.
If harmful DNA mutations are detected early, then an oncologist can treat the cancer early.
Early detection is a major determinant for surviving cancer.
In the case of lung cancer, doctors know that patients with lung cancer often have mutations in the middle of a gene called EGFR (epidermal growth factor receptor).
If a biopsy is performed on the patient’s lung tissue then a doctor can test the DNA in the lung tumor cells. This DNA test will determine whether the EGFR gene is mutated.
Fortunately, targeted drugs (Tagrisso, Iressa, etc.) are available that specifically treat lung cancer patients that test positive for an EGFR mutation.
Performing a tumor tissue biopsy and sequencing the DNA within the cancer cells allows doctors to identify patients who are most likely to benefit from a targeted therapy.
These tumor tissue DNA tests are often performed with a technique called whole exome sequencing (described above). This DNA test only examines a small portion of the entire 3 billion base human genome. Whole exome sequencing of tumor tissue will search for hundreds of cancer related DNA mutations within the gene regions of our genome.
Genetic mutations and cancer
Detecting mutations in these genes can help diagnose the related cancer. Targeted therapies exist for cancers with these biomarkers.
| Mutated gene | Related cancer |
|---|---|
| ALK | lung cancer (NSCLC) |
| EGFR | lung cancer (NSCLC) |
| BRAF | melanoma |
| BCR-ABL | leukemia (CML) |
| BRCA | ovarian cancer |
| IDH2 | leukemia (AML) |
Visit this resource to explore various targeted therapies currently available for cancer treatment.
Sidenote: A genetic test performed on tumor tissue may reveal a cancer-related DNA mutation that was inherited from the parents. If 50% of the alleles examined in the biopsy contain a cancer-related DNA mutation, then it is possible the mutation was passed on from a parent (a germline mutation). If only a small percent of the cells in a biopsy contain the cancer-related DNA mutation, then the mutation likely arose during the lifetime of the individual (somatic mutation). If you learn you carry a cancer-related DNA mutation that is also germline, you should discuss with a genetic counselor or other health provider as this information may have implications for other family members (1).
The tumor tissue biopsies described above are considered invasive because they involve a physician removing body tissue from the cancer location. In contrast, a liquid biopsy is a non-invasive variation of a tumor tissue biopsy.
How does a liquid biopsy work?
As a cancer is progressing, some tumor cells will die and, inadvertently, release their DNA into the patient’s bloodstream. Therefore, cancer patients, even early cancer patients, have fragments of cancer cell DNA floating freely in their bloodstream. This is called cell-free, tumor cell DNA.
By taking a blood sample and isolating out the cell-free, tumor cell DNA, a specialist can then sequence this DNA and search for DNA mutations that are associated with a particular from of cancer. Once armed with this knowledge, the oncologist may be able to prescribe a targeted therapy that delivers a higher chance of survival.
Cell-free liquid biopsies are performed on blood as well as urine and saliva. Multiple companies have recently received FDA approval for cell-free DNA liquid biopsy tests.
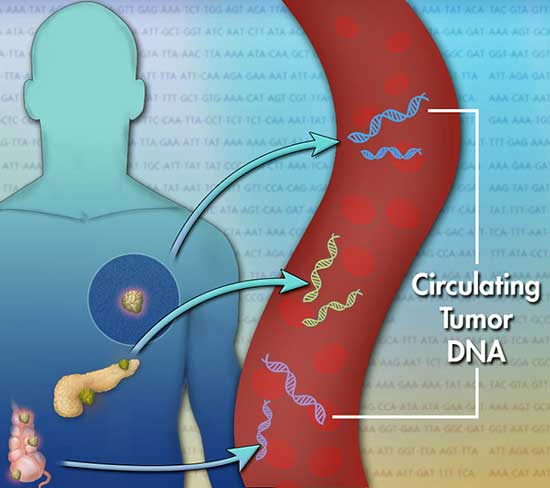
image: J. Bailey: CC BY-SA 4.0
Dying tumor cells release small pieces of DNA into the patient’s bloodstream.
A liquid biopsy will collect this cell-free DNA and search for specific DNA mutations.
Sidenote: At this time, it seems more work needs to be done before we can rely on liquid biopsies (cell free DNA) to accurately diagnose cancers (1). Stay tuned…
Genetic testing before pregnancy
Some couples decide to have carrier screening before they have children.
Carrier screening is a type of genetic test that can tell you whether you carry a gene for a certain genetic disorder.
If a couple performs a carrier screen before pregnancy, they can find out the chances of having a child with a genetic disorder.
Every child will inherit 1 copy of their father’s genome and 1 copy of their mother’s genome. These 2 sources of DNA combine to create a unique human genome for that new individual. This process also explains why we all carry 2 versions of the same gene (1 version from each parent).
We know that many genetic diseases are heritable (passed on from parents to children). If the prospective parents are aware of their genetic background, they may be able to take steps to reduce the chance their child will be born with a genetic abnormality.
What is a carrier?
A carrier is a person who has 1 bad gene for a genetic disorder. Carriers usually don’t show symptoms for the disease, often because the 1 good version of the gene is sufficient to perform the biological task.
In the absence of symptoms, a carrier may not know that they carry the problematic gene. This situation could lead to a carrier inadvertently passing the bad gene on to their child. If both parents pass on 1 bad version of a gene, then the child may inherit 2 bad versions of the same gene (1 from each parent). That scenario can cause a genetic disease.
Genetic diseases are passed on to children in different ways. Some genetic diseases are Mendelian, in which case they are caused by either recessive or dominant genes.
Cystic fibrosis (CF) is an example of a recessive genetic disorder. CF causes persistent lung infections and limits the ability to breathe. CF appears when a child inherits 2 bad versions of the CFTR gene.
As with other recessive disorders, a child must inherit 2 bad copies of the CFTR gene (1 from each parent) in order to exhibit symptoms of cystic fibrosis.
Each time two CTFR carriers have a child, the chances are:
- 25 percent their child will have CF
- 50 percent the child will be a CFTR carrier but will not have CF
- 25 percent the child will not be a carrier of the CFTR gene and will not have CF
Approximately one in every 31 Americans is a symptom-less carrier of 1 bad version of the CFTR gene.
Historical sidenote: Gregor Mendel, an Austrian monk, taught us these probabilities for recessive disorders in 1866. Mendel discovered the probability of inheritance by examining the color and shape of pea plants. The scientific community largely ignored Mendel’s discovery for the next 40 years. In 1905, Mendel’s work was re-examined and it soon became the cornerstone of a new field of science, called genetics.

Gregor Mendel
Fortunately, scientists have gathered good information regarding the heritability of many genetic disorders. The National Institute of Health provides this resource to educate the general public about these diseases. Once you select a genetic disease, scroll down to the inheritance section to learn how this disease is passed on from parent to child.
Inheritance sidenote: It should be noted that many genetic disorders do not follow the simple math of Mendelian inheritance. Often a disease is passed on to a child via other mechanisms such as incomplete dominance, codominance, epigenetics, sex-linked traits and contributions from more than one gene. In these situations, the probability of the child inheriting a genetic disease becomes less certain.
How do couples initiate carrier screening?
Typically, the partner who is more at risk will be tested first. For example, if one partner has a history of genetic disease in their family then it makes sense for that partner to be tested first.
If test results reveal that the first tested partner is not a carrier, then no additional testing is needed for recessive disorders. If test results show that the first partner is a carrier, then the other partner should be tested.
Speak with your primary care physician to receive advice on carrier screening.
If a couple receives carrier screen results and finds the risk of passing on a genetic disease is high, they may want to consider alternative options.
Below, I describe 4 possible options.
- IVF (in vitro fertilization): With this option, both the man and woman contribute to the child’s DNA. The woman’s egg is fertilized with the man’s sperm outside of the body, in a clinic. The fertilized eggs (embryos) can be screened for chromosomal abnormalities and certain genetic disorders. IVF also allows the couple to choose the sex of the child. Once tested, the healthy embryos are implanted into the woman’s body and brought to term. IVF costs about 20-25K and insurance companies are currently hesitant to pay for this procedure. See footnote below.
- Sperm or egg donor: This option involves only 1 parent contributing to the child’s DNA. In general, the sperm donor scenario (in utero insemination) is a much more affordable option than using an egg donor (IVF with another woman’s egg.)
- Have the child: Once a couple determines there’s a reasonable risk their offspring will be born with a genetic disorder, they may still decide to have a child and hope the inheritance pattern works in their favor. In this case, the couple can consider prenatal testing early in the pregnancy (CVS, NIPT). An early diagnosis with prenatal testing may allow for early treatment. See section below.
- Adoption: If there is a significant risk of passing on a genetic disorder, a couple may decide to adopt.
IVF can be combined with genetic screening. This process is known as preimplantation genetic screening (PGT). PGT is becoming increasingly more common. This makes sense considering we are currently learning more about genetic mutations that can cause or correlate with genetic disorders. The CDC recently reported that 22% of IVF procedures now involve genetic screening via PGT. In the future, I imagine most IVFs will be accompanied by PGT screening. As mentioned 20-25K is a lot to pay for this service, however, the alternative could be significantly more costly. If parents produce a child with a genetic disorder, the costs of managing and treating that child will quickly exceed 25K. Furthermore, if a gene therapy is the best possible treatment option for that child, then that one-time gene therapy treatment may cost 2-5 million. How will insurance payers deal with a 5 million pricetag on a gene therapy? In my opinion, they will respond by initiating coverage for the full cost of PGT and IVF for eligible parents.
Genetic testing options during pregnancy
Ok, let’s say you’re pregnant. You and your partner have created a new embryo. Congrats!
The embryo received 1 copy of the father’s genome and 1 copy of the mother’s genome. All of this DNA recombined in a completely new manner to create a unique individual.
As a couple, you now have an excellent opportunity to examine the genetic health of your new embryo.
Prenatal diagnosis allow parents to diagnose the health of their child before the child is born.
A skilled physician can remove a small amount of biological material from a pregnant mother. Using this material, a clinic can screen for genetic abnormalities and other developmental issues in the unborn baby.
This information has tremendous value. The sooner the parents are aware of a health problem, the sooner any corrective actions can be made. Some health problems can be treated before the baby is born, others conditions will greatly improve if the baby is treated immediately after childbirth.
artwork: Ryan Kitson
Three different tests are available to screen for genetic abnormalities in your unborn child.
First, I list the main points for each test. I then discuss in more details below.
Chorionic villus sampling (CVS)
- usually performed between 10 and 12 weeks
- determines sex of child, tests for genetic disorders
- catheter inserts into uterus and removes placenta material
- invasive procedure, small chance of miscarriage
- performed between the 15th and 20th week of pregnancy or later
- determines sex of child, tests for genetic disorders and other issues
- needle inserted into womb, amniotic fluid removed
- invasive procedure, small chance of miscarriage
Noninvasive prenatal test with cell-free DNA (NIPT with cfDNA)
- can be performed as early as 10 weeks
- determines sex of child, tests for aneuploidy (chromosomal defects)
- DNA from fetus collected from mother’s blood sample
- noninvasive procedure, no chance of miscarriage
Chorionic villus sampling (CVS)
- Pronounced: CORE-ree-on-ik VILL-us SAM-pling
CVS is a prenatal test in which a small amount of chorionic villi is removed from the placenta and analyzed for genetic disorders. Chorionic villi are wispy projections of placenta. The placenta is an organ that connects a developing fetus with the mother’s uterus. Placenta tissue develops from the embryo, so when you analyze placenta tissue, you’re examining the new cells of the fetus.
This procedure is usually performed between week 10 and 13 of pregnancy. To extract chorionic villi, a physician will insert a catheter (thin tube) through the vagina, through the cervix and into the uterus. An ultrasound helps guide the careful removal of a small amount of placenta material. CVS was first performed in 1983 by Giuseppe Simoni, an Italian biologist.
Similar to amniocentesis, CVS yields fetal cells. Each one of these cells contains the genome (all the genetic material) of the new fetus. This DNA can be analyzed for the same genetic abnormalities that would be detected by amniocentesis. However, since CVS does not yield amniotic fluid, there are certain health issues that will be missed, including: Rh incompatibility, neural tube defects and other birth defects not detected via genetic analysis. Therefore, a couple may opt for amniocentesis if they previously had a child with a neural tube defect or blood incompatibility issue.
The clear advantage to CVS is that it’s performed about 5 weeks earlier than amniocentesis.
It should be noted that both CVS and amniocentesis are invasive procedures. Depending on the type of CVS performed, a CVS procedure may involve slightly more risk of injury to the fetus.
Amniocentesis
- pronounced: AM-nee-oh-sen-TEE-sis
A fetus floats in amniotic fluid while it’s developing in the womb. During this time, some cells sluff off the developing fetus and become suspended in the amniotic fluid. During the 15th week of pregnancy, a physician can insert a needle through the mother’s abdominal wall, into the uterus and extract about 20 milliliters of this amniotic fluid.
Both the amniotic fluid and the floating fetal cells are analyzed in a laboratory. An amniocentesis test can reveal if the fetus is experiencing an infection, improper lung development, Rh blood incompatibility or a neural tube defect.
In addition, DNA from the fetal cells can be analyzed to determine the sex of the child and a handful of chromosomal abnormalities, including Down syndrome (trisomy 21) Edwards syndrome (trisomy 18) and Turner syndrome (monosomy X).
DNA from these fetal cells can also be analyzed with either whole exome sequencing or a broad panel screen for genetic mutations. Both of these tests would search for DNA mutations that could affect the health of the child.

artwork: Ryan Kitson
Sidenote: In the absence of a medical condition (a phenotype), there is currently little value to using whole exome sequencing on a fetus. Sequencing all the genes of a fetus will yield lots of DNA sequence variation…but without a condition to correlate with this variation, it will be difficult to make sense of the information (1). Progress is being made in this regard, explore this BabySeq page to learn more.
Amniocentesis was first developed by Robert Lisle Gadd and presented to the public in 1965. The groundbreaking procedure is now performed with the aid of an ultrasound. Ultrasound allows physicians to visualize the fetus and avoid its injury during the procedure. Unfortunately, amniocentesis also involves the risk of fluids leaking from the amniotic sac after the procedure. This activity could contribute to a miscarriage.
Noninvasive prenatal testing (NIPT with cfDNA)
Recent advances in DNA testing have allowed a new form of noninvasive prenatal testing. DNA from the developing fetus can be isolated from a blood sample that is collected from a pregnant women. Clinicians can then test this fetal DNA for genetic abnormalities. This procedure is called noninvasive prenatal testing (NIPT) using cell‐free DNA (cfDNA). It became available in 2011 and is now offered widely throughout the world.
By simply drawing blood from a pregnant women, the genetic material of her fetus can be analyzed and screened for certain genetic disorders.
In general, NIPT with cfDNA screens for:
- sex of the fetus
- fetal rheusus blood type
- chromosomal disorders: Down syndrome, Edwards syndrome, Patau syndrome, etc.
- some single gene disorders
How is it possible to collect fetal DNA from the blood of a pregnant women?
It turns out that DNA material is released from cells of the fetus as a natural process of embryo development. Much of this cell-free DNA then circulates in the mother’s blood.
Scientists determined that between 3-6% of all the cell-free DNA circulating in a pregnant women is actually DNA from the fetus. In 1997, Lo and colleagues published a report in Lancet demonstrating that this fetal DNA can be captured from the woman’s blood and analyzed with PCR to search for genetic disorders.
The striking benefit to this procedure is that it’s completely safe for both mother and child. There is a zero chance of miscarriage associated with this NIPT procedure.
A pregnant women may visit a clinic to have blood drawn as early as 10 weeks into pregnancy.

artwork: Ryan Kitson
NIPT with cfDNA is currently a popular testing option for women over 35 and for couples with a previous history of producing children with chromosomal disorders. The test is considered very accurate for detecting chromosomal abnormalities.
NIPT has been endorsed as a valuable test by professional organizations from several nations, however, it should be noted that NIPT is not considered diagnostic. If a NIPT test finds evidence for a genetic disorder, a couple may want to confirm results with an invasive test (CVS or amniocentesis). In addition, post-test counseling is highly recommended to help parents navigate the test results.
NIPT sidenote: At this time, NIPT with cfDNA is used commercially to detect chromosomal abnormalities (called aneuploidies). The NIPT test is not commonly used commercially to screen for a panel of genetic mutations. As our ability to distinguish between maternal and fetal DNA improves, we may see a panel of genetic mutations included with NIPT in the future. As it stands now (2018), NIPT screening for a panel of genetic mutations exists in limited contexts. For example, Baylor University in Texas currently offers PreSeek, a commercial product offered by a lab in a University.
Please consult with a genetic counselor to determine the optimal test for your specific situation. This resource helps you find a genetic counselor.
Screening newborns for genetic disorders
Every 8 seconds a baby is born in the United States.
The vast majority of these babies are born healthy, however, some arrive with a serious medical condition.
Approximately 3% of babies are born with some form of birth defect (cleft palate, spina bifida, down syndrome, heart valve defects, etc.)
If the medical condition is genetic, the birth defect may have been anticipated with pre-natal screening (CVS, amniocentesis, NIPT).
When your child is first born you will want to perform a newborn screen for genetic disorders.
Most newborn screens are performed 12 to 48 hours after the baby is born, usually before the child has left the hospital.
If a baby does have a serious medical condition, the parents should be aware of the problem as soon as possible. Prompt identification and management of a medical condition can sometimes prevent any serious complications associated with the disease.

artwork: Ryan Kitson
Currently, all 50 states in the United States require newborn screening programs.
Nearly every child born in the United States is screened for genetic abnormalities shortly after birth.
This procedure is often called a heel stick. During a heel stick, a trained hospital employee pricks the heel of the newborn and collects drops of blood on a small piece of filter paper. This blood is then subjected to biochemical and possibly DNA sequence analysis in order to identify the presence of genetic abnormalities. For example, a newborn screen will test for cystic fibrosis, sickle cell anemia, phenylketonuria, SCID and galactosemia.
The cost of newborn screening is approximately $160.00 and this cost should be easily covered by Medicare or private insurance. In theory, the only reason a child in the US would not receive newborn genetic screening is if the parents refused the test.
As of 2019, a minimum of 35 genetic disorders are screened as standard of care for newborns. Surprisingly, each state in the US screens for a different panel of genetic disorders. When assembling a list of screened disorders, each state considers its own laws, financing and the state’s incidence rate for each specific disorder.
This state by state approach creates discrepancies in the genetic disorders screened across the country.
For example, California currently screens for 61 genetic disorders, while West Virginia only screens for 38 disorders.
Follow this link to learn which genetic disorders are screened in your state.
The future of newborn screening…
As we discover more treatments for genetic disorders, the list of screened genetic disorders should continue to expand in each state.
Spinal muscular atrophy (SMA) is a potentially lethal genetic disorder caused by a DNA mutation in a gene called SMN1. Normally, this SMN1 gene codes for a motor neuron protein that allows muscles to operate correctly. In the absence of a proper gene sequence for SMN1, a newborns’ muscle cells don’t function correctly. Tragically, 1 in every 6,000 babies are born with SMA.
In July of 2018, SMA was added to the Recommended Uniform Screening Panel (RUSP) for newborn babies in the United States. The Secretary of Health, Alex Azar, made this decision partly because a treatment for SMA does now exist. A drug named Spinraza (developed by Biogen) was approved by the FDA in late 2016.
A newborn genetic screen for SMA will go a long way towards ensuring an afflicted child receives treatment as soon as possible.
How do we treat genetic disorders?
A child born with a genetic disorder (monogenic) is missing the accurate DNA sequence for one particular gene.
The human body is programmed by approximately 20,000 genes. These genes provide instructions to build a human body and keep it operational throughout a human lifespan.
In a healthy newborn, each of these genes provides instructions to build a corresponding protein at the right time and right place in the developing body. The proteins perform biological work in the body, this work keeps the body functional.
If a newborn is missing a gene, then certain organs and tissues in their body will be lacking a protein. That can be a problem…
Imagine popping the hood of a car and randomly removing one component to the engine.
Would the car still run?
Maybe…maybe not.
It depends what’s missing. If the windshield wiper fluid is removed, the windshield will accumulate dirt but the car will still work.
If the brake fluid is removed then we’ve got a problem.
Missing certain genes is disastrous while missing other genes is manageable.
It depends on the gene.
Before decoding the human genome in 2003, we only knew the genetic basis for a handful of genetic disorders.
We now know the genetic basis for more than 6,000 genetic disorders.
Categories of treatment options for genetic disorders
Simply because we understand the genetic basis for a disorder does not mean a simple treatment exists.
The research and medical community has been working hard to develop various treatment options.
Here, I list categories of treatments for genetic disorders.
- surgery – heart defect surgery, bone marrow transplant
- life support device – ventilator, kidney dialysis, feeding tube
- small molecule drugs – pharmaceuticals
- large biologic drugs – monoclonal antibodies that target misshapen proteins
- RNA therapy – RNAi or exon skipping technology (Exondys 51, Spinraza)
- specialized diets – metabolism errors can be treated with diet, baby formula, etc.
- gene therapy – introduce missing gene back into the correct cells (Luxturna)
Below, I discuss 2 of these treatment options in more detail: gene therapy and specialized diets for inborn errors of metabolism.
Inborn Errors of Metabolism
All genetic disorders are a challenge for both the newborn and the parents. However, if diagnosed early, some genetic disorders can be managed without surgery or elaborate medication. Genetic disorders that are classified as inborn errors of metabolism (IEM) can often be treated with a specific diet or with supplements.
What are Inborn Errors of Metabolism?
Our body is constantly breaking apart large complex molecules, we call this metabolism. We metabolize our food so that the nutrients in the food can be absorbed into our body.
Unfortunately, some people are born missing one piece of their metabolic machinery.
Enzymes are a class of proteins responsible for breaking apart the complex molecules in our body. These enzymes do the heavy lifting when it comes to metabolism. The human genome contains thousands of genes that encode for enzyme proteins. Each of these enzymes is responsible for metabolizing a specific molecule in our body.
If a newborn is missing a gene for a metabolic enzyme, then they are afflicted with an Inborn Error of Metabolism.
artwork: Ryan Kitson
PKU is an example of an inborn error of metabolism that can be addressed with a restricted diet.
Phenylketonuria, or PKU, is a rare inherited disorder that prevents the proper digestion of a nutrient called phenylalanine. Phenylalanine is an amino acid that is found in most protein-based foods.Normally, the PKU gene encodes for an enzyme that metabolizes phenylalanine. In the absence of a PKU gene, a newborn accumulates toxic levels of phenylalanine in their body, eventually this build up results in damage to the nerve cells in their brain.
Fortunately, PKU is one of the genetic disorders included in the newborn screen for all US states.
The treatment for PKU is a low protein diet. A special formula is available for babies who test positive for PKU. This restricted diet should begin immediately after birth.
Please visit this page to learn more information about genetic disorders that can be treated with diet or supplementation.
Gene Therapy
To truly fix, or cure, a genetic disorder, you need to insert a fully functional gene into the genome of the patient who is missing that gene.
If this is done correctly, the patient will produce the correct protein in the correct cells and the symptoms of the disease will greatly diminish.
We call this approach gene therapy.
Scientists have been working hard to develop a safe and effective form of gene therapy for the past 40 years.
Fortunately, we are beginning to see the fruits of their labor.
The first gene therapy for a genetic disorder was approved in December of 2017. A company named Spark received FDA approval for a therapy to treat a rare eye disorder that causes blindness.
Got 1 minute?
This video does a great job explaining the basics of gene therapy. Want more details? I cover the topic extensively here.
Below I discuss some of the genetic disorders that are currently being pursued by gene therapy companies.
Inherited retinal disease
Almost all children and adults living with inherited retinal disease (IRD) will eventually go blind. The disorder is caused by a mutation in the RPE65 gene. Spark, a gene therapy company, uses an adeno-associated virus (AAV) to deliver a functional RPE65 gene into the retinal cells of IRD patients.
The list price for this treatment began at $425K per eye. For those keeping score at home, that’s $850K for both eyes.
To learn more details about the pricing for Luxturna, I recommend reading this discussion of fair cost for gene therapies.
Hemophilia
Hemophilia is a genetic disease characterized by an inability to produce a blood clotting factor.
Hemophiliacs suffer extensive bleeding events throughout their life. Sometimes, even bumping a leg against a table is sufficient trauma to cause a dangerous bleed.
In the case of hemophilia A, individuals are missing a gene called Factor VII. A few different companies are currently in competition to develop the first FDA approved gene therapy that can replace the Factor VII gene in hemophiliac patients.
Spark, BioMarin and Sangamo all have viable candidates in their product pipeline. Each company is progressing through clinical trials to determine the efficacy and safety of their hemophilia gene therapy.
Spinal Muscular Atrophy
Spinal muscular atrophy (SMA) is a rare, inherited neuro-muscular disorder. SMA patients harbor defects in their SMN1 gene. The absence of functional SMN1 protein causes major problems in motor neuron cells, which can lead to death at a young age.
A company named AveXis developed a gene therapy for SMA. In April 2018, Novartis announced they will purchase AveXis for $8.7 billion in cash. Novartis is now moving this therapy, called Zolgensma, through clinical trials and analysts expect an FDA approval in 2019.
There is a lot of interest in the price tag for Zolgensma. Novartis grabbed headlines in November of 2018 by stating that their “one-time, potentially curative” therapy would be cost-effective at a list price of $4-5 million.
In contrast, the healthcare industry watchdog named ICER has issued a report stating that they think Zolgensma could be cost-effective at a one-time price of $2 million. ICER acknowledged we need more data regarding the long term durability and health benefits of the gene therapy before they can effectively calculate a cost-effective price.
Either way, expect a big number for Zolgensma in the near future.
Looking to the future…
As of 2019, forward progress is being made for many other genetic disorders: Pompe disease, cystic fibrosis, sickle cell anemia, etc.
Click here to see my list of gene therapies currently moving through clinical trials.
I will update this page every 6 months to keep up with gene therapy advancements.
As you can tell, I have great interest in this subject. It’s a thrill to be alive during this Genomic Age.
By applying our knowledge of genetics toward medicine and biotech, we stand to greatly reduce human suffering worldwide.
If you think others will benefit from information on this web page, please share with an icon below.
Thanks for reading.
~ Dr. Kevin Curran
References
- Personal communications with genetic counselor, Andrea Procko
- Mukherjee, Siddhartha. The gene: An intimate history. Simon and Schuster, 2017.
- Mukherjee, Siddhartha. The emperor of all maladies: a biography of cancer. Simon and Schuster, 2010.
- Evans, James P., Bradford C. Powell, and Jonathan S. Berg. “Finding the rare pathogenic variants in a human genome.” Jama 317.18 (2017): 1904-1905.
- Minear, Mollie A., et al. “Noninvasive prenatal genetic testing: current and emerging ethical, legal, and social issues.” Annual review of genomics and human genetics 16 (2015): 369-398.
- Lo, YM Dennis, et al. “Quantitative analysis of fetal DNA in maternal plasma and serum: implications for noninvasive prenatal diagnosis.” The American Journal of Human Genetics62.4 (1998): 768-775.
- Lo, YM Dennis, et al. “Presence of fetal DNA in maternal plasma and serum.” The lancet 350.9076 (1997): 485-487.
- Mayo Clinic, PKU
- NIH, genetic disorders
- NIH, newborn screen
- NIH, rare disease
- NIH, actionable genes
- NIH, spinal muscular atrophy
- CDC, birth defects
- Baby first test, newborn screen
- Baylor, preseek
- DNA testing, Antonio Regalado
- Cystic Fibrosis, carrier test
- BabySeq Project
Artwork
Photos of Alberto Giacometti sculptures were taken by the author at the Guggenheim Museum in NYC. The Guggenheim graciously allowed their use on this page.
Plastic sculptures designed by Ryan Kitson.